Structure and content of RAVmodel
Sehyun Oh
July 27, 2022
Source:vignettes/Contents.Rmd
Contents.RmdSetup
Install and load package
if (!require("BiocManager"))
install.packages("BiocManager")
BiocManager::install("GenomicSuperSignature")Download RAVmodel
You can download GenomicSuperSignature from Google Cloud bucket using
GenomicSuperSignature::getModel function. Currently
available models are built from top 20 PCs of 536 studies (containing
44,890 samples) containing 13,934 common genes from each of 536 study’s
top 90% varying genes based on their study-level standard deviation.
There are two versions of this RAVmodel annotated with different gene
sets for GSEA: MSigDB C2 (C2) and three priors from PLIER
package (PLIERpriors). In this vignette, we are showing the
C2 annotated model.
Note that the first interactive run of this code, you will be asked
to allow R to create a cache directory. The model file will be stored
there and subsequent calls to getModel will read from the
cache.
RAVmodel <- getModel("C2", load=TRUE)
#> [1] "downloading"Content of RAVmodel
RAVindex is a matrix containing genes in rows and RAVs
in columns. colData slot provides the information on each
RAVs, such as GSEA annotation and studies involved in each cluster.
metadata slot stores model construction information.
trainingData slot contains the information on individual
studies in training dataset, such as MeSH terms assigned to each
study.
RAVmodel
#> class: PCAGenomicSignatures
#> dim: 13934 4764
#> metadata(8): cluster size ... version geneSets
#> assays(1): RAVindex
#> rownames(13934): CASKIN1 DDX3Y ... CTC-457E21.9 AC007966.1
#> rowData names(0):
#> colnames(4764): RAV1 RAV2 ... RAV4763 RAV4764
#> colData names(4): RAV studies silhouetteWidth gsea
#> trainingData(2): PCAsummary MeSH
#> trainingData names(536): DRP000987 SRP059172 ... SRP164913 SRP188526
version(RAVmodel)
#> [1] "1.1.1"
geneSets(RAVmodel)
#> [1] "MSigDB C2 version 7.1"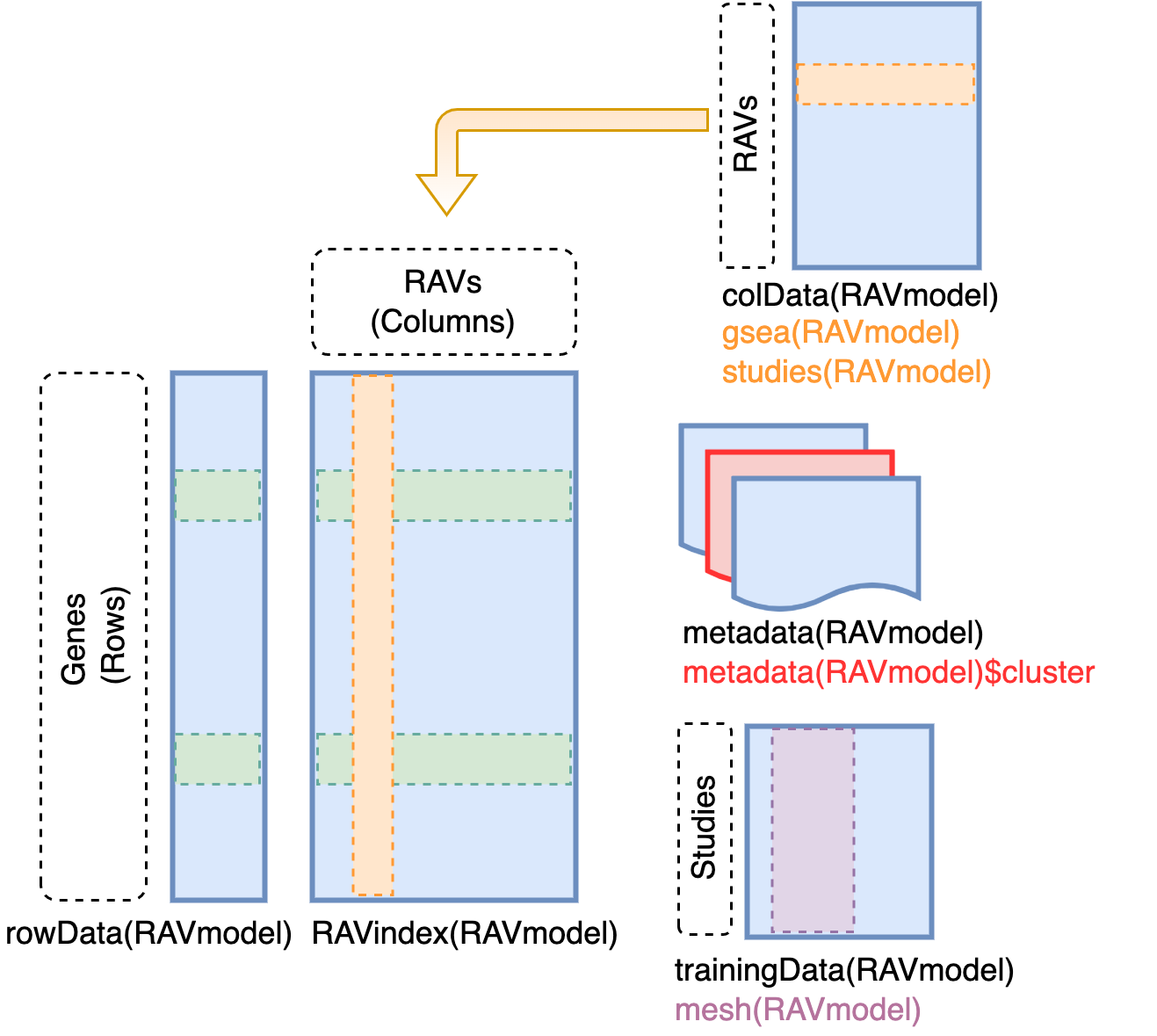
RAVindex
Replicable Axis of Variation (RAV) index
is the main component of GenomicSuperSignature. It serves as an index
connecting new datasets and the existing database. You can access it
through GenomicSuperSignature::RAVindex (equivalent of
SummarizedExperiment::assay). Rows are genes and columns
are RAVs.
Here, RAVmodel consists of 13,934 genes and 4,764 RAVs.
class(RAVindex(RAVmodel))
#> [1] "matrix" "array"
dim(RAVindex(RAVmodel))
#> [1] 13934 4764
RAVindex(RAVmodel)[1:4, 1:4]
#> RAV1 RAV2 RAV3 RAV4
#> CASKIN1 0.002449532 -2.845471e-03 0.001601715 -0.009366156
#> DDX3Y 0.006093438 1.870528e-03 -0.004346019 -0.006094526
#> MDGA1 0.009999975 2.610302e-03 -0.002831447 -0.006710757
#> EIF1AY 0.007923652 8.507021e-05 0.002322275 -0.007850784Metadata for RAVmodel
Metadata slot of RAVmodel contains information related to the model building.
names(metadata(RAVmodel))
#> [1] "cluster" "size" "k" "n" "MeSH_freq"
#> [6] "updateNote" "version" "geneSets"-
cluster: cluster membership of each PCs from the training dataset
-
size: an integer vector with the length of clusters, containing the number of PCs in each cluster
-
k: the number of all clusters in the given RAVmodel
-
n: the number of top PCs kept from each study in the training dataset -
geneSets: the name of gene sets used for GSEA annotation
-
MeSH_freq: the frequency of MeSH terms associated with the training dataset. MeSH terms like ‘Humans’ and ‘RNA-seq’ are top ranked (which is very expected) because the training dataset of this model is Human RNA sequencing data. -
updateNote: a brief note on the given model’s specification -
version: the version of the given model
head(metadata(RAVmodel)$cluster)
#> DRP000987.PC1 DRP000987.PC2 DRP000987.PC3 DRP000987.PC4 DRP000987.PC5
#> 1 2 3 4 5
#> DRP000987.PC6
#> 6
head(metadata(RAVmodel)$size)
#> RAV1 RAV2 RAV3 RAV4 RAV5 RAV6
#> 6 21 4 7 3 3
metadata(RAVmodel)$k
#> [1] 4764
metadata(RAVmodel)$n
#> [1] 20
geneSets(RAVmodel)
#> [1] "MSigDB C2 version 7.1"
head(metadata(RAVmodel)$MeSH_freq)
#>
#> Humans RNA-Seq Transcriptome
#> 1163 591 576
#> RNA Whole Exome Sequencing Sequencing, RNA
#> 372 284 277
updateNote(RAVmodel)
#> [1] "GSEA with set.seed"
metadata(RAVmodel)$version
#> [1] "1.1.1"Studies in each RAV
You can find which studies are in each cluster using
studies method. Output is a list with the length of
clusters, where each element is a character vector containing the name
of studies in each cluster.
length(studies(RAVmodel))
#> [1] 4764
studies(RAVmodel)[1:3]
#> $Cl4764_01
#> [1] "DRP000987" "SRP059172" "SRP059959" "SRP118741" "SRP144178" "SRP183186"
#>
#> $Cl4764_02
#> [1] "DRP000987" "ERP022839" "ERP104463" "ERP106996" "ERP109789" "ERP114435"
#> [7] "SRP059039" "SRP059172" "SRP065317" "SRP071547" "SRP073061" "SRP074274"
#> [13] "SRP075449" "SRP092158" "SRP096736" "SRP107337" "SRP111402" "SRP125008"
#> [19] "SRP133442" "SRP135684" "SRP155483"
#>
#> $Cl4764_03
#> [1] "DRP000987" "SRP001540" "SRP056840" "SRP125396"You can check which PC from different studies are in RAVs using
PCinRAV.
PCinRAV(RAVmodel, 2)
#> [1] "DRP000987.PC2" "ERP022839.PC2" "ERP104463.PC1" "ERP106996.PC2"
#> [5] "ERP109789.PC2" "ERP114435.PC1" "SRP059039.PC3" "SRP059172.PC2"
#> [9] "SRP065317.PC1" "SRP071547.PC3" "SRP073061.PC2" "SRP074274.PC5"
#> [13] "SRP075449.PC2" "SRP092158.PC2" "SRP096736.PC2" "SRP107337.PC3"
#> [17] "SRP111402.PC1" "SRP125008.PC2" "SRP133442.PC1" "SRP135684.PC4"
#> [21] "SRP155483.PC2"Silhouette width for each RAV
Silhouette width ranges from -1 to 1 for each cluster. Typically, it
is interpreted as follows:
- Values close to 1 suggest that the observation is well matched to the
assigned cluster
- Values close to 0 suggest that the observation is borderline matched
between two clusters
- Values close to -1 suggest that the observations may be assigned to
the wrong cluster
For RAVmodel, the average silhouette width of each cluster is a quality control measure and suggested as a secondary reference to choose proper RAVs, following validation score.
x <- silhouetteWidth(RAVmodel)
head(x) # average silhouette width of the first 6 RAVs
#> 1 2 3 4 5 6
#> -0.05470163 0.06426256 -0.01800335 -0.04005584 0.05786189 -0.02520973GSEA on each RAV
Pre-processed GSEA results on each RAV are stored in RAVmodel and can
be accessed through gsea function.
class(gsea(RAVmodel))
#> [1] "list"
class(gsea(RAVmodel)[[1]])
#> [1] "data.frame"
length(gsea(RAVmodel))
#> [1] 4764
gsea(RAVmodel)[1]
#> $RAV1
#> Description NES
#> REACTOME_MITOCHONDRIAL_TRANSLATION REACTOME_MITOCHONDRIAL_TRANSLATION 2.503359
#> REACTOME_TRANSLATION REACTOME_TRANSLATION 2.271857
#> CROONQUIST_STROMAL_STIMULATION_UP CROONQUIST_STROMAL_STIMULATION_UP -3.634893
#> pvalue qvalues
#> REACTOME_MITOCHONDRIAL_TRANSLATION 1.00000e-10 9.218794e-08
#> REACTOME_TRANSLATION 1.00000e-10 9.218794e-08
#> CROONQUIST_STROMAL_STIMULATION_UP 1.20798e-10 9.218794e-08MeSH terms for each study
You can find MeSH terms associated with each study using
mesh method. Output is a list with the length of studies
used for training. Each element of this output list is a data frame
containing the assigned MeSH terms and the detail of them. The last
column bagOfWords is the frequency of the MeSH term in the
whole training dataset.
length(mesh(RAVmodel))
#> [1] 536
mesh(RAVmodel)[1]
#> $DRP000987
#> score identifier concept name
#> 35822 1000 DRP000987 C0042333 Genetic Variation
#> 58627 18575 DRP000987 C0017431 Genotype
#> 149067 37162 DRP000987 C0021247 Indonesia
#> 216848 74145 DRP000987 C0030498 Parasites
#> 716101 22634 DRP000987 C4760607 RNA-Seq
#> 761253 55509 DRP000987 C0024530 Malaria
#> 832289 96999 DRP000987 C0024535 Malaria, Falciparum
#> 1116527 161422 DRP000987 C0032150 Plasmodium falciparum
#> 1230230 7197 DRP000987 C0020969 Innate Immune Response
#> 1468242 162422 DRP000987 C0003062 Animals
#> 1573542 19938 DRP000987 C0035668 RNA
#> 1596889 162422 DRP000987 C0086418 Humans
#> 1627781 36315 DRP000987 C0752248 Transcriptome Analysis
#> 1689323 57176 DRP000987 C3178810 Transcriptome
#> explanation
#> 35822 Forced Non-Leaf Node Lookup:genetic variations
#> 58627 Forced Lookup:genotyping
#> 149067 RtM via: Indonesia, RtM via: Indonesians, Forced Leaf Node Lookup:indonesians
#> 216848 RtM via: Parasites, Forced Leaf Node Lookup:parasites, checkForceMH Boosted
#> 716101 RtM via: RNA-Seq, Forced New Lookup:rna-seq
#> 761253 RtM via: Malaria, Forced Non-Leaf Node Lookup:malaria
#> 832289 RtM via: Plasmodium falciparum infection
#> 1116527 RtM via: Plasmodium falciparum, Forced Leaf Node Lookup:plasmodium falciparums, checkForceMH Boosted
#> 1230230 Entry Term Replacement for "Immunity, Innate", RtM via: Immunity, Innate, Forced Non-Leaf Node Lookup:innate immune responses
#> 1468242 Parasites, CT Treecode Lookup: B01.050.500.714 (Parasites)
#> 1573542 RtM via: RNA
#> 1596889 Forced Leaf Node Lookup:humans, CT Text Lookup: human, CT Text Lookup: humans, CT Text Lookup: patient, CT Text Lookup: patients, Forced Leaf Node Lookup:humans, CT Text Lookup: human, CT Text Lookup: humans, CT Text Lookup: patient, Parasites, CT Treecode Lookup: B01.050.500.714 (Parasites)
#> 1627781 Entry Term Replacement for "Gene Expression Profiling", RtM via: Gene Expression Profiling, Forced Non-Leaf Node Lookup:transcriptome analysis
#> 1689323 RtM via: Transcriptome, Forced Leaf Node Lookup:transcriptomes
#> bagOfWords
#> 35822 11
#> 58627 20
#> 149067 2
#> 216848 7
#> 716101 591
#> 761253 6
#> 832289 4
#> 1116527 5
#> 1230230 8
#> 1468242 209
#> 1573542 372
#> 1596889 1163
#> 1627781 66
#> 1689323 576PCA summary for each study
PCA summary of each study can be accessed through
PCAsummary method. Output is a list with the length of
studies, where each element is a matrix containing PCA summary results:
standard deviation (SD), variance explained by each PC (Variance), and
the cumulative variance explained (Cumulative).
length(PCAsummary(RAVmodel))
#> [1] 536
PCAsummary(RAVmodel)[1]
#> $DRP000987
#> DRP000987.PC1 DRP000987.PC2 DRP000987.PC3 DRP000987.PC4
#> SD 42.6382804 21.97962707 12.94848367 11.89265640
#> Variance 0.3580557 0.09514629 0.03302091 0.02785537
#> Cumulative 0.3580557 0.45320196 0.48622287 0.51407824
#> DRP000987.PC5 DRP000987.PC6 DRP000987.PC7 DRP000987.PC8
#> SD 10.89460616 10.53655583 9.47278967 8.97268098
#> Variance 0.02337622 0.02186495 0.01767287 0.01585607
#> Cumulative 0.53745446 0.55931942 0.57699228 0.59284836
#> DRP000987.PC9 DRP000987.PC10 DRP000987.PC11 DRP000987.PC12
#> SD 8.43652010 7.90810771 7.60865968 7.076401846
#> Variance 0.01401774 0.01231676 0.01140165 0.009862254
#> Cumulative 0.60686609 0.61918285 0.63058449 0.640446748
#> DRP000987.PC13 DRP000987.PC14 DRP000987.PC15 DRP000987.PC16
#> SD 6.89881271 6.766783788 6.703417100 6.6042884
#> Variance 0.00937346 0.009018116 0.008850009 0.0085902
#> Cumulative 0.64982021 0.658838324 0.667688332 0.6762785
#> DRP000987.PC17 DRP000987.PC18 DRP000987.PC19 DRP000987.PC20
#> SD 6.44257163 6.245257583 6.042236748 5.992461574
#> Variance 0.00817466 0.007681604 0.007190294 0.007072317
#> Cumulative 0.68445319 0.692134796 0.699325091 0.706397408Session Info
sessionInfo()
#> R version 4.2.0 (2022-04-22)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Ubuntu 20.04.4 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/liblapack.so.3
#>
#> locale:
#> [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
#> [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
#> [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
#> [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
#> [9] LC_ADDRESS=C LC_TELEPHONE=C
#> [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#>
#> attached base packages:
#> [1] stats4 stats graphics grDevices utils datasets methods
#> [8] base
#>
#> other attached packages:
#> [1] GenomicSuperSignature_1.4.0 SummarizedExperiment_1.26.1
#> [3] Biobase_2.56.0 GenomicRanges_1.48.0
#> [5] GenomeInfoDb_1.32.2 IRanges_2.30.0
#> [7] S4Vectors_0.34.0 BiocGenerics_0.42.0
#> [9] MatrixGenerics_1.8.1 matrixStats_0.62.0
#> [11] BiocStyle_2.24.0
#>
#> loaded via a namespace (and not attached):
#> [1] bitops_1.0-7 fs_1.5.2 bit64_4.0.5
#> [4] filelock_1.0.2 httr_1.4.3 doParallel_1.0.17
#> [7] RColorBrewer_1.1-3 rprojroot_2.0.3 tools_4.2.0
#> [10] backports_1.4.1 bslib_0.4.0 utf8_1.2.2
#> [13] R6_2.5.1 DBI_1.1.3 colorspace_2.0-3
#> [16] GetoptLong_1.0.5 tidyselect_1.1.2 curl_4.3.2
#> [19] bit_4.0.4 compiler_4.2.0 textshaping_0.3.6
#> [22] cli_3.3.0 desc_1.4.1 DelayedArray_0.22.0
#> [25] bookdown_0.27 sass_0.4.2 scales_1.2.0
#> [28] rappdirs_0.3.3 pkgdown_2.0.6 systemfonts_1.0.4
#> [31] stringr_1.4.0 digest_0.6.29 rmarkdown_2.14
#> [34] XVector_0.36.0 pkgconfig_2.0.3 htmltools_0.5.3
#> [37] dbplyr_2.2.1 fastmap_1.1.0 rlang_1.0.4
#> [40] GlobalOptions_0.1.2 RSQLite_2.2.15 shape_1.4.6
#> [43] jquerylib_0.1.4 generics_0.1.3 jsonlite_1.8.0
#> [46] dplyr_1.0.9 car_3.1-0 RCurl_1.98-1.7
#> [49] magrittr_2.0.3 GenomeInfoDbData_1.2.8 Matrix_1.4-1
#> [52] Rcpp_1.0.9 munsell_0.5.0 fansi_1.0.3
#> [55] abind_1.4-5 lifecycle_1.0.1 stringi_1.7.8
#> [58] yaml_2.3.5 carData_3.0-5 zlibbioc_1.42.0
#> [61] BiocFileCache_2.4.0 blob_1.2.3 grid_4.2.0
#> [64] parallel_4.2.0 crayon_1.5.1 lattice_0.20-45
#> [67] circlize_0.4.15 knitr_1.39 ComplexHeatmap_2.12.0
#> [70] pillar_1.8.0 ggpubr_0.4.0 rjson_0.2.21
#> [73] ggsignif_0.6.3 codetools_0.2-18 glue_1.6.2
#> [76] evaluate_0.15 BiocManager_1.30.18 png_0.1-7
#> [79] vctrs_0.4.1 foreach_1.5.2 gtable_0.3.0
#> [82] purrr_0.3.4 tidyr_1.2.0 clue_0.3-61
#> [85] assertthat_0.2.1 cachem_1.0.6 ggplot2_3.3.6
#> [88] xfun_0.31 broom_1.0.0 rstatix_0.7.0
#> [91] ragg_1.2.2 tibble_3.1.8 iterators_1.0.14
#> [94] memoise_2.0.1 cluster_2.1.3 ellipsis_0.3.2