Research
Public Data Reuse
Database Search and Transfer Learning
We developed a method for interpreting new transcriptomic datasets through instant comparison to public datasets without high-performance computing requirements. This method consists of a pre-computed model and an out-of-the-box Bioconductor software package, GenomicSuperSignature, which applies the model to new data. Using human datasets, we demonstrated our method’s efficient and coherent database search capabilities, robustness to batch effects and heterogeneous training data, and transfer learning capacity. We are currently expanding this method to different species (e.g., mouse, yeast) and different omics data types (e.g., scRNAseq, metagenomics). Additionally, leveraging the transfer learning capacity of this method, we are developing a systematic framework for inferring various phenotypes from transcriptomic data.
Cross-study Metadata Standardization

To enhance the FAIRness of public omics data, we aim to systematically harmonize and standardize metadata across major public biomedical data repositories. We are performing large-scale manual metadata curation for publicly available omics data while building an automated metadata harmonization pipeline that leverages various Natural Language Processing (NLP) techniques, including Large Language Models (LLMs), using manually harmonized metadata as a gold standard. The standardization process integrates ontologies, a data reconciliation system, and scientific provenance tracking to ensure the interpretability, accuracy, and reliability of the harmonized data. Currently, we are targeting several major public omics data repositories for standardization. The outcome of this project will transform how researchers discover and integrate public data into new research topics, ultimately accelerating biological discovery.
Comprehensive Cancer Data Analysis
Histopathology Image Data Analysis
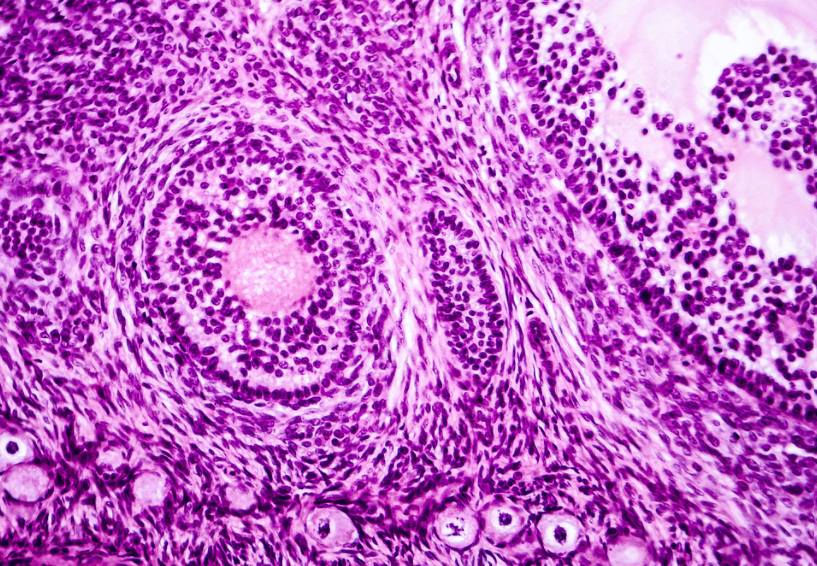
Histopathological images provide unparalleled insights into tissue architecture, cellular morphology, and tumor spatial organization. However, analyzing these images often requires non-R software, unlike other high-throughput omics data and downstream statistical analyses that are frequently performed in the R/Bioconductor environment. A seamless connection between these platforms is critical for integrative multi-modal analyses that combine both omics and image data. To address this gap, we are creating standardized workflows that process raw image files and extract image features accessible in R, while also developing a comprehensive repository of image features extracted from The Cancer Genome Atlas (TCGA) images.
Microbiome
Antibiotics
Antibiotics are among the most widely prescribed drugs in the United States, and adverse reactions to them represent significant morbidity. While the mechanisms of action for different classes of antibiotics are well understood at the molecular level, these reactions occur within the complex and individualized ecosystem of the human microbiome, where our understanding is much more limited. Our goal is to construct a curated and harmonized database of antibiotic-exposed microbiome data and use this resource to understand both the personalized effects of antibiotic treatments on the gut microbiome and the development of antibiotic resistance. This knowledge will ultimately help maximize the benefits of antibiotics while minimizing adverse effects.
Supporting Interdisciplinary Research
Cloud Computing
Advancements in sequencing technologies and new data collection methods are producing unprecedented volumes of biological data. However, the computational infrastructure and technical skills required to leverage these vast quantities of biological data make such analyses challenging for basic, translational, and clinical researchers. As a part of NHGRI’s Genomic Analysis, Visualization and Informatics Lab-space (AnVIL) project, we are developing a user-friendly working environment that enables non-technical researchers to utilize public datasets and Cloud-implemented workflows.