---
title: "My Awesome Report"
author: "Alison Goff"
date: ""
output: html_document
---Session 5_2. Reproducible projects with Rmarkdown
Questions
- What is R Markdown?
- How can I integrate my R code with text and plots?
- How can I convert .Rmd files to .html?
Learning Objectives
- Create a .Rmd document containing R code, text, and plots
- Create a YAML header to control output
- Understand basic syntax of (R)Markdown
- Customize code chunks to control formatting
Introduction to R Markdown
R Markdown is a flexible type of document that allows you to seamlessly combine executable R code, and its output, with text in a single document. These documents can be readily converted to multiple static and dynamic output formats, including PDF (.pdf), Word (.docx), and HTML (.html).
Creating an R Markdown file
To create a new R Markdown document in RStudio, click File -> New File -> R Markdown:
Then click on ‘Create Empty Document’. Normally you could enter the title of your document, your name (Author), and select the type of output, but we will be learning how to start from a blank document.
Basic components of R Markdown
To control the output, a YAML header is needed:
The header is defined by the three hyphens at the beginning (---) and the three hyphens at the end (---). In this header, the only required field is the output:, which specifies the type of output you want. This can be an html_document, a pdf_document, or a word_document. We will use an HTML document for now.
The rest of the fields can be deleted, if you don’t need them. After the header, to begin the body of the document, you start typing after the end of the YAML header (i.e. after the second ---).
YAML is a human-readable data serialization language that is often used for writing configuration files. Depending on whom you ask, YAML stands for yet another markup language or YAML ain’t markup language (a recursive acronym), which emphasizes that YAML is for data, not documents.
YAML files are simpler to read as they use indentation to determine the structure and indicate nesting. Tab characters are not allowed by design, to maintain portability across systems, so white-spaces (i.e., literal space characters) are used instead.
Comments can be identified with a pound or hash symbol (#). It’s always a best practice to use comments, as they describe the intention of the code. YAML does not support multi-line comment, each line needs to be suffixed with the pound character.
Markdown syntax
Markdown is a popular markup language that allows you to add formatting elements to text, such as bold, italics, and code.
Headers
A # in front of text indicates to Markdown that this text is a heading. Adding more #s make the heading smaller, i.e. one # is a first level heading, two ##s is a second level heading, etc. upto the 6th level heading.
# Title
## Section
### Sub-section
#### Sub-sub section
##### Sub-sub-sub section
###### Sub-sub-sub-sub sectionMarkup languages are computer languages that are used to structure, format, or define relationships between different parts of text documents with the help of symbols or tags inserted in the document.
Bold and Italics
- You can make things bold by surrounding the word with double asterisks,
**bold**, or double underscores,__bold__ - You can make things italicize using single asterisks,
*italics*, or single underscores,_italics_. - You can also combine bold and italics to write something really important with triple-asterisks,
***really***, or underscores,___really___or a combination of asterisks and underscores,**_really_**,_**really**_.
More
To create code-type font, surround the word with backticks, `code-type`. You can also create a list for the variables (using -, +, * keys), an ordered list (using numbers), and nested items (using tab-indenting). For more Markdown syntax see the following reference guide.
Rendering
You can render the document into HTML by clicking the Knit button in the top of the Source pane (top left). The knit function takes an input file, extracts the R code in it according to a list of patterns, evaluates the code and writes the output in another file. If you haven’t saved the document yet, you will be prompted to do so when you knit for the first time.
Writing an R Markdown report
You need to load both packages and data within your R markdown document - it is not enough to load packages and data from the console. To load these, we will need to create a ‘code’ chunk’ at the top of our document (below the YAML header).
A code chunk can be inserted by clicking Code > Insert Chunk, or by using the keyboard shortcuts Ctrl+Alt+I on Windows and Linux, and Cmd+Option+I on Mac.
The syntax of a code chunk is:
```{r chunk-name}
"Here is where you place the R code that you want to run."
```An R Markdown document knows that this text is not part of the report from the three backticks, ```, that begins and ends the chunk. It also knows that the code inside of the chunk is R code from the r inside of the curly braces ({}). After the r you can add a name for the code chunk . Naming a chunk is optional, but recommended. Each chunk name must be unique, and only contain alphanumeric characters and -.
To load packages (e.g., tidyverse) and the surveys data table (from session3_2), we will insert a chunk and call it ‘setup’. Since we don’t want this code or the output to show in our knitted HTML document, we add an include = FALSE option after the code chunk name ({r setup, include = FALSE}).
The file paths you give in a .Rmd document, e.g. to load a .csv file, are relative to the .Rmd document, not the project root.
Insert table
When you add/modify your code chunks in you rmarkdown file, you don’t need to knit the whole document. Instead, you can run the code chunk with the green triangle in the top right corner of the the chunk.
surveys %>%
filter(!is.na(weight), # remove missing weight
!is.na(hindfoot_length), # remove missing hindfoot_length
!is.na(sex)) %>% # remove missing sex
group_by(plot_type) %>%
summarize(plots = paste(unique(plot_id), collapse = ",")) %>%
knitr::kable(col.names = c("Plot Type", "Plot Number")) # format nicely| Plot Type | Plot Number |
|---|---|
| Control | 2,17,12,11,22,14,4,8 |
| Long-term Krat Exclosure | 3,15,19,21 |
| Rodent Exclosure | 5,24,10,16,23,7 |
| Short-term Krat Exclosure | 18,20,6,13 |
| Spectab exclosure | 1,9 |
To make the table in our output document formatted nicely, we can use the kable() function from the knitr package. The kable() function takes the output of your R code and knits it into a nice looking HTML table. You can also specify different aspects of the table, e.g. the column names, a caption, etc.
Many different R packages can be used to generate tables. Some of the more commonly used options are listed in the table below.
| Name | Creator(s) | Description |
|---|---|---|
| condformat | Oller Moreno (2022) | Apply and visualize conditional formatting to data frames in R. It renders a data frame with cells formatted according to criteria defined by rules, using a tidy evaluation syntax. |
| DT | Xie et al. (2023) | Data objects in R can be rendered as HTML tables using the JavaScript library ‘DataTables’ (typically via R Markdown or Shiny). The ‘DataTables’ library has been included in this R package. |
| formattable | Ren and Russell (2021) | Provides functions to create formattable vectors and data frames. ‘Formattable’ vectors are printed with text formatting, and formattable data frames are printed with multiple types of formatting in HTML to improve the readability of data presented in tabular form rendered on web pages. |
| flextable | Gohel and Skintzos (2023) | Use a grammar for creating and customizing pretty tables. The following formats are supported: ‘HTML’, ‘PDF’, ‘RTF’, ‘Microsoft Word’, ‘Microsoft PowerPoint’ and R ‘Grid Graphics’. ‘R Markdown’, ‘Quarto’, and the package ‘officer’ can be used to produce the result files. |
| gt | Iannone et al. (2022) | Build display tables from tabular data with an easy-to-use set of functions. With its progressive approach, we can construct display tables with cohesive table parts. Table values can be formatted using any of the included formatting functions. |
| huxtable | Hugh-Jones (2022) | Creates styled tables for data presentation. Export to HTML, LaTeX, RTF, ‘Word’, ‘Excel’, and ‘PowerPoint’. Simple, modern interface to manipulate borders, size, position, captions, colours, text styles and number formatting. |
| pander | Daróczi and Tsegelskyi (2022) | Contains some functions catching all messages, ‘stdout’ and other useful information while evaluating R code and other helpers to return user specified text elements (e.g., header, paragraph, table, image, lists etc.) in ‘pandoc’ markdown or several types of R objects similarly automatically transformed to markdown format. |
| pixiedust | Nutter and Kretch (2021) | ‘pixiedust’ provides tidy data frames with a programming interface intended to be similar to ’ggplot2’s system of layers with fine-tuned control over each cell of the table. |
| reactable | Lin et al. (2023) | Interactive data tables for R, based on the ‘React Table’ JavaScript library. Provides an HTML widget that can be used in ‘R Markdown’ or ‘Quarto’ documents, ‘Shiny’ applications, or viewed from an R console. |
| rhandsontable | Owen et al. (2021) | An R interface to the ‘Handsontable’ JavaScript library, which is a minimalist Excel-like data grid editor. |
| stargazer | Hlavac (2022) | Produces LaTeX code, HTML/CSS code and ASCII text for well-formatted tables that hold regression analysis results from several models side-by-side, as well as summary statistics. |
| tables | Murdoch (2022) | Computes and displays complex tables of summary statistics. Output may be in LaTeX, HTML, plain text, or an R matrix for further processing. |
| tangram | Garbett et al. (2023) | Provides an extensible formula system to quickly and easily create production quality tables. The processing steps are a formula parser, statistical content generation from data defined by a formula, and rendering into a table. |
| xtable | Dahl et al. (2019) | Coerce data to LaTeX and HTML tables. |
| ztable | Moon (2021) | Makes zebra-striped tables (tables with alternating row colors) in LaTeX and HTML formats easily from a data.frame, matrix, lm, aov, anova, glm, coxph, nls, fitdistr, mytable and cbind.mytable objects. |
Customizing chunk output
We mentioned using include = FALSE in a code chunk above. There are additional options available to customize how the code chunks are presented in the output document. The full R Markdown code chunk option can be found here. Below are some of the widely used options:
| Option | Options | Output |
|---|---|---|
eval |
TRUE or FALSE
|
Whether or not the code within the code chunk should be run. |
echo |
TRUE or FALSE
|
Choose if you want to show your code chunk in the output document. echo = TRUE will show the code chunk. |
include |
TRUE or FALSE
|
Choose if the output of a code chunk should be included in the document. FALSE means that your code will run, but will not show up in the document. |
warning |
TRUE or FALSE
|
Whether or not you want your output document to display potential warning messages produced by your code. |
message |
TRUE or FALSE
|
Whether or not you want your output document to display potential messages produced by your code. |
fig.align |
default, left, right, center
|
Where the figure from your R code chunk should be output on the page. |
Note that the default settings for the above chunk options are all TRUE
Insert plots
We are using the murders data table from the dslabs package.
library(dslabs)
library(ggthemes)
library(ggrepel)
r <- murders |>
summarize(rate = sum(total) / sum(population) * 10^6) |>
pull(rate)
plots <- murders |> ggplot(aes(population/10^6, total, label = abb)) +
geom_abline(intercept = log10(r), lty = 2, color = "darkgrey") +
geom_point(aes(col=region), size = 3) +
geom_text_repel() +
scale_x_log10() +
scale_y_log10() +
xlab("Populations in millions (log scale)") +
ylab("Total number of murders (log scale)") +
ggtitle("US Gun Murders in 2010") +
scale_color_discrete(name = "Region") +
theme_economist()
plots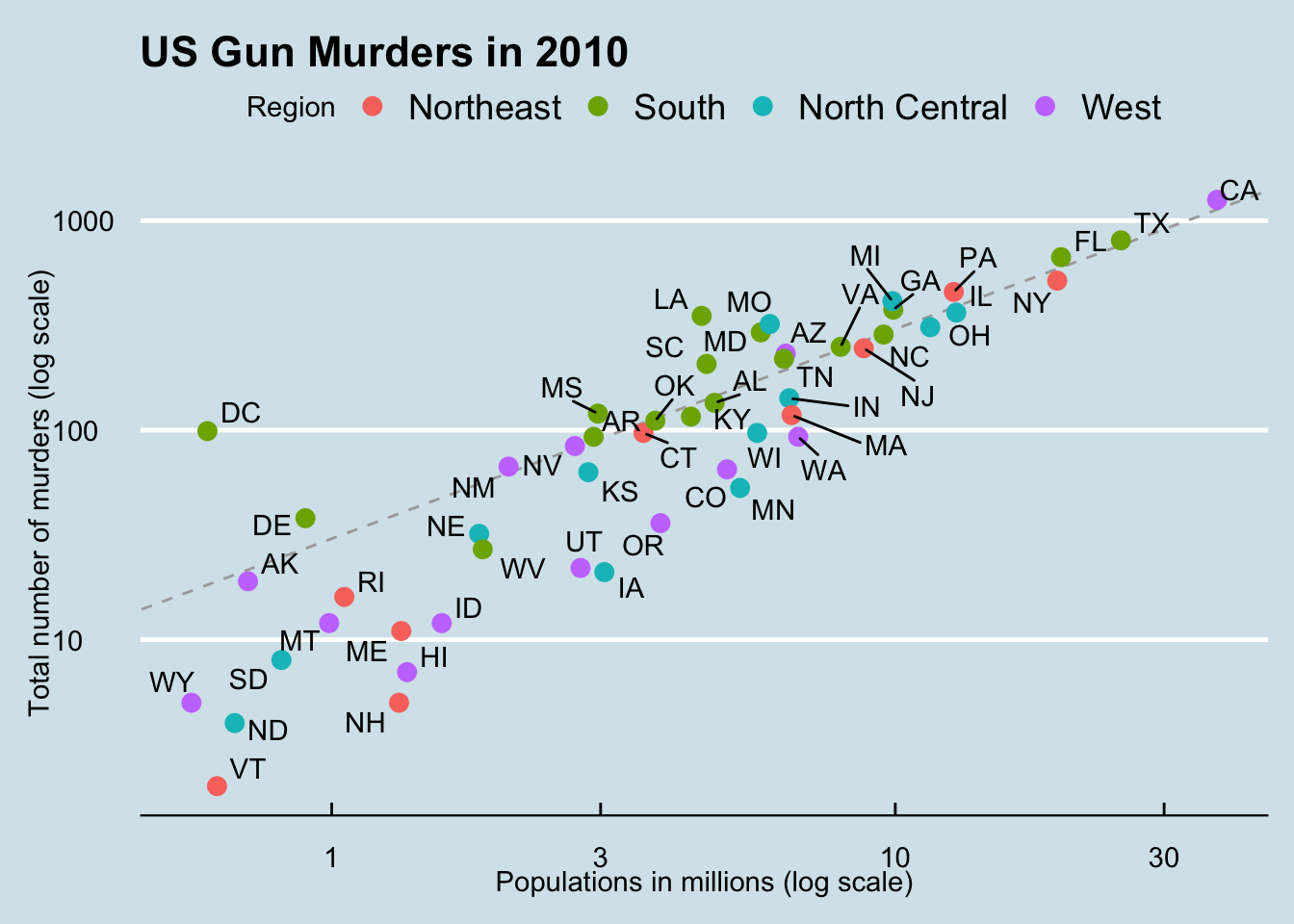
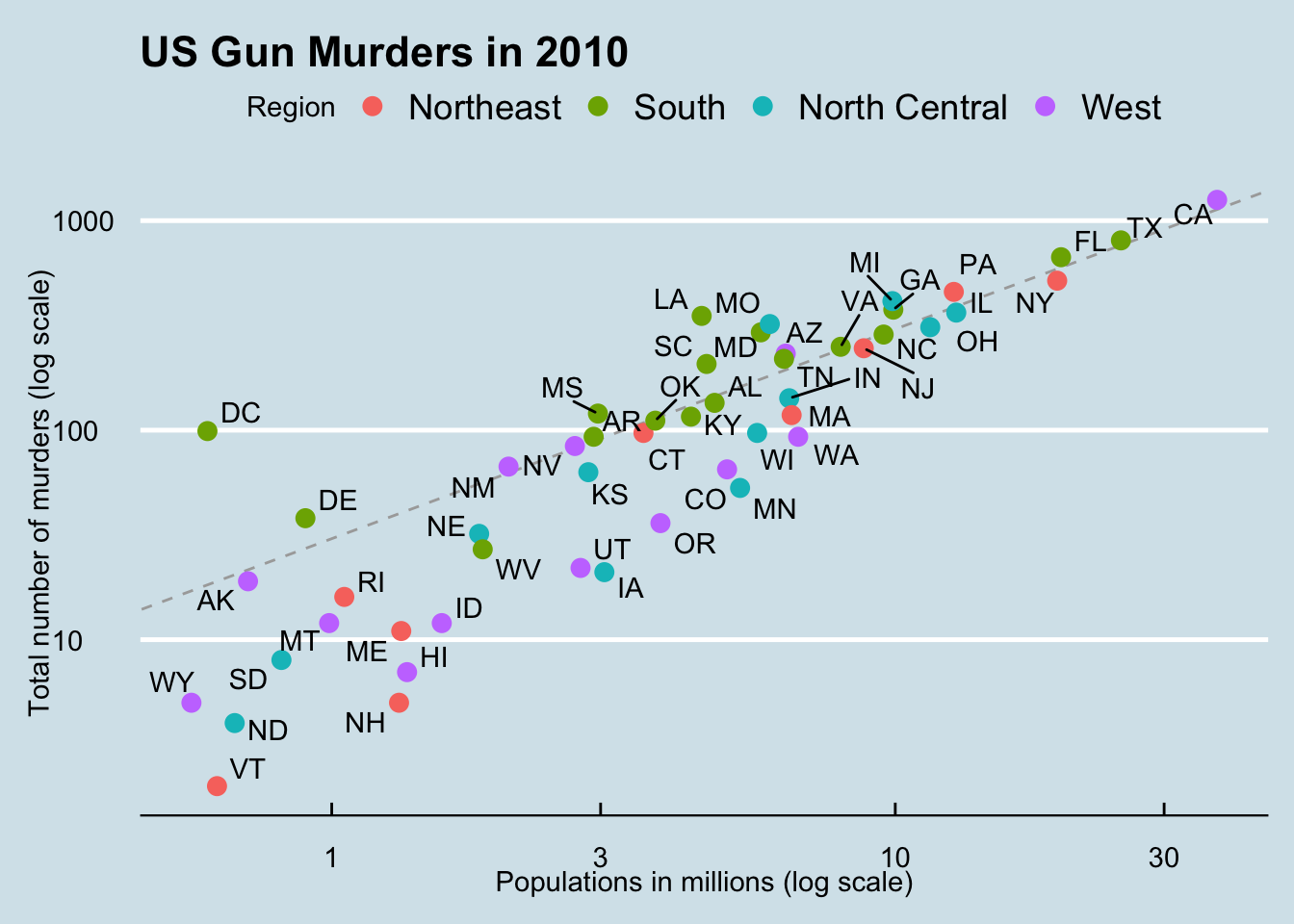
We can use R Markdown chunk options. For example, we can add a caption with the chunk option fig.cap and resize the plot size using out.width and out.height:
```{r fig.cap = "Figure 1. Summary", out.width="60%", out.height="60%"}
plots
```
References
https://datacarpentry.org/r-socialsci/06-rmarkdown.html
http://rafalab.dfci.harvard.edu/dsbook-part-1/dataviz/ggplot2.html